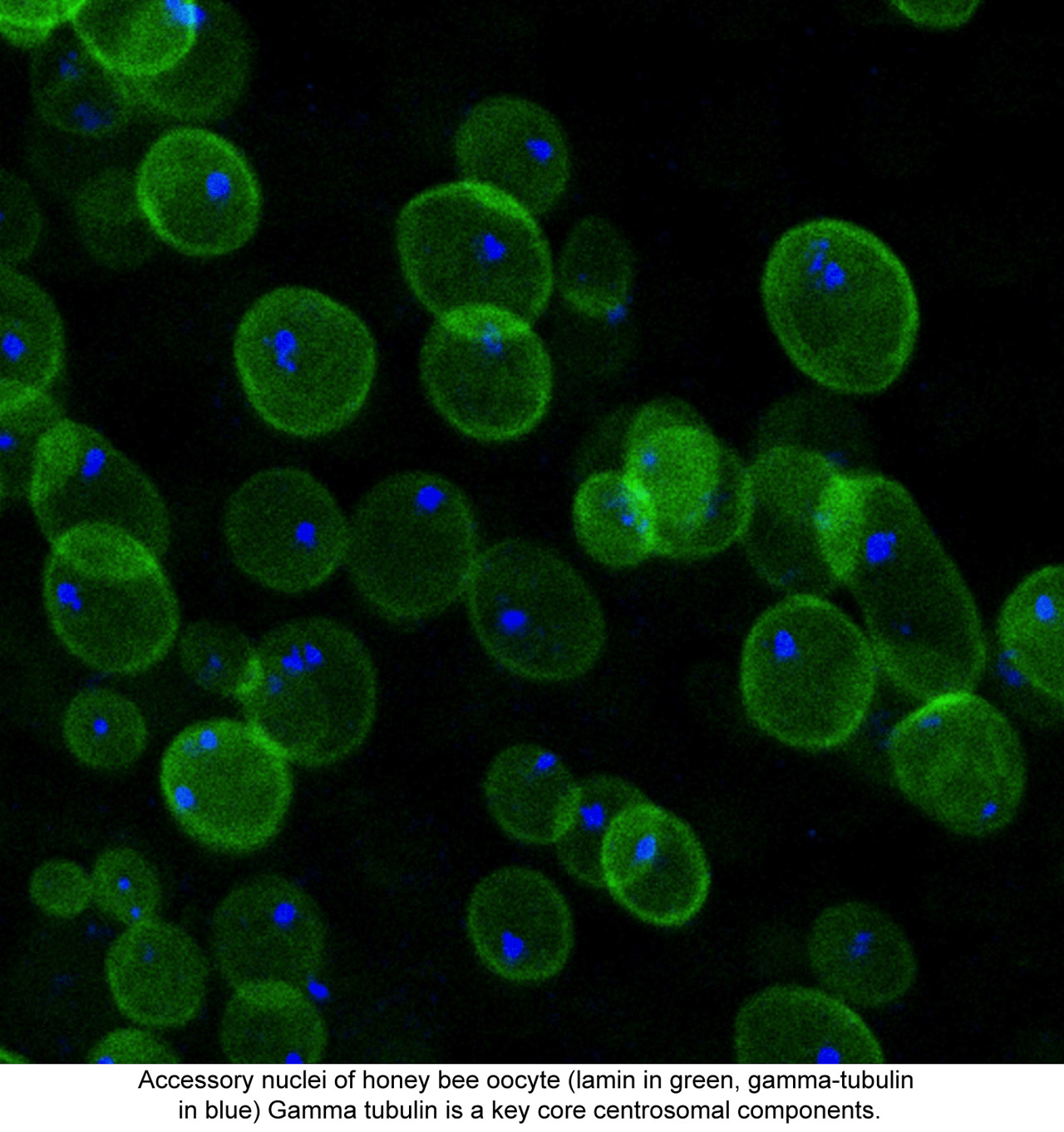
The UCSC Life Sciences Microscopy Center serves the biological research community at UCSC and provides personalized assistance on various aspects of imaging, from experimental design to training on the shared microscopes and image analysis.
Light Microscopy
UCSC’s Life Sciences Microscopy Center is a shared use facility that provides imaging instrumentation for advanced biomedical research. The center is located in Sinsheimer labs and the Biomedical Science Buildings and is supported by the UCSC Division of Physical and Biological Sciences and the California Institute for Quantitative Biosciences (QB3).
Electron Microscopy
The electron microscope is a user-sponsored resource supervised by Professor Melissa Jurica. Please direct inquiries to facilities director Benjamin Abrams. Inquiries related to the cryo-electron microscopy facility can be directed to Vitor Hugo Balasco Serrao. Additional information can be found in the Biomolecular Cryo-electron Microscopy Facility website.
Fee Schedule
| FY24 Rates | UCSC hourly rate |
UCSC 24 Hour Period |
External, Non-Profit hourly rate |
External, Non-Profit 24 Hour Period |
External, For-Profit hourly rate |
External, For-Profit 24 Hour Period |
|---|---|---|---|---|---|---|
| Zeiss AxioZoom | $9.33 | $144.96 | $14.46 | $224.69 | $28.92 | $449.38 |
| Leica DM5500B Widefield | $9.33 | – | $14.46 | – | $28.92 | – |
| Keyence Widefield | $7.73 | – | $11.98 | – | $23.96 | – |
| Zeiss Live Cell Imaging Station | $9.33 | $74.64 | $14.46 | $115.69 | $28.92 | $231.38 |
| Zeiss AxioImager | $9.33 | $144.96 | $14.46 | $224.69 | $28.92 | $449.38 |
| Solamere Spinning Disk Confocal | $15 | $240 | $23.25 | $372.00 | $46.50 | $744.00 |
| Leica SP5 Confocal | $26.52 | $424.36 | $41.11 | $657.76 | $82.21 | $1,315.52 |
| Zeiss 880 Confocal | $29.18 | $466.80 | $45.23 | $723.54 | $90.46 | $1,447.08 |
| Technical Assistance | $51.84 | – | $80.35 † | – | $160.70 † | – |
| TRAINING: Recharge Scope | $0* | – | $80.35 † | – | $160.70 † | – |
| TRAINING: Non-Recharge Scope | $51.84 | – | $80.35 | – | $160.70 | – |
| ADD-ON: Assisted Imaging | $51.84 | – | $80.35 | – | $160.70 | |
| ADD-ON: On-Scope Assay Development | $51.84 | – | $80.35 | – | $160.70 | – |
Please Note: All ADD-ON rates are IN ADDITION to the standard scope rate. Scope access for External Users is dependent upon schedule availability; please contact the facility director to learn more.
*UCSC Users receiving training on a non-recharge scope will not incur a training fee, but may incur a scope use fee for the duration of their training session; if training must occur from their own account
†Non-UCSC Users receiving training on a non-recharge scope will incur a scope use fee for the duration of their training session
Instrument Details
The UCSC Life Sciences Microscopy Center manages several widefield and confocal microscopes available for use by the UCSC research community. Some are owned by the CIRM-funded UCSC Institute for the Biology of Stem Cells, and others belong to individual laboratories but are administered by the center.
Zeiss SteREO Discovery V20 Teaching Dissection Scope
Best suited for direct observation of specimens that require a large amount (up to 110mm) of working distance (wd) and for teaching applications where simultaneous observation of specimen or procedure by student and teacher is beneficial.
Stand: dual observation head with MARC remote control
Stage: motorized with focus and height control.
Transmitted light modes: direct and oblique illumination with directional or diffuse reflectors
Reflected light illumination: two variable mount LED
Magnification range: 4.7x – 94.5x
Ownership: common
Location: 319 Sinsheimer Labs
Zeiss Axiozoom Microscope
Best suited for samples normally viewed under a dissections scope and requiring large working distances, but where higher magnification is needed. Well suited for thin (less than 15 microns) samples mounted on a standard microscope slide with a coverslip. Non-labeled and fluorescently-labeled samples may be used.
Stand: Axiozoom V.16
Stage: motorized stage with transmitted light
Fluorescent filters available:
DAPI: Fset49, EX 365 : DC 396 : EM BP 445/50
GFP: Fset38, EX BP 470/40 : DC 495 : EM BP 525/50
Cy3: Fset43, EX BP 550/25 : DC 570 : EM BP 605/70
Cy5: Fset50, EX BP 640/30 : DC 660 : EM BP 690/50
Transmitted light modes: bright-field, bright-field plus, relief contrast, dark-field
Objectives:
0.5x/0.125 NA (wd – 114mm)
1x/.025 NA (wd- 56mm)
2.3x/0.57 NA (wd-10.6mm)
Software: Zeiss Zen with Z-stack, tiling and stitching modules
Camera: Zeiss AxioCam HRm (monochrome)
Ownership: lab-owned
Location: Sinsheimer 312C
Leica Widefield Microscope
Best suited for thin (less than 10 microns) samples mounted on a standard microscope slide with a coverslip. Non-labeled, fluorescently-labeled, and colorimetric-dye-labeled samples may be used.
Stand: upright, Leica DM5500B
Stage: motorized stage for 3D stack, tiling, and multi-point image acquisition.
Fluorescent filters available:
DAPI: 116000232 EX BP 360/40 : DC 400 : EM BP 470/40
GFP: 11532366 EX BP 470/40 : DC 500 : EM BP 525/50
Texas Red: 11513885 EX 560/40 : DC 595 : EM BP 570/75
Cy5: 11600230 EX BP 620/60 : DC 660 : EM BP 625/75
Cy3: 11600231 EX 545/40 : DC 565 : EM 610/75 (note: Cy3 cube is not normally on the scope but may be exchanged in upon request)
Transmitted light modes: bright-field, DIC, and polarized light
Objectives:
4x/0.1 NA (wd-18mm)
10x/0.3 NA (wd-11mm)
20x/0.5 NA (DIC) (wd-1.15mm)
40x/0.75 NA (DIC) (wd- 0.40mm)
63x/ 0.6-1.4 NA oil (DIC) (wd-0.1mm)
100x/0.7-1.4 NA oil (DIC) (wd-0.09mm)
Software: Leica Application Suite Advanced Fluorescence V2.6.9.7266
Camera(s): Leica DCF360 monochrome, Leica DFC450 color
Ownership: common
Location: 111 Sinsheimer Labs
Keyence Biorevo BZ-9000 Digital Widefield Microscope
Best suited for thin (less than 10 microns) samples mounted on a standard microscope slide with a coverslip, but can also accommodate samples in a small petri dish. Non-labeled, fluorescently-labeled, and colorimetric-dye-labeled samples may be used. Especially good for applications requiring image tiling or for samples that have large fluctuations along the Z-axis.
Stand: inverted, Keyence BZ-9000 stand-alone unit
Stage: motorized stage for 3D stack, tiling, and multipoint image acquisition; digital navigation for eyepiece-free use
Fluorescent filters available:
DAPI: EX BP 360/40 : DC 400 : EM BP 460/50
GFP: EX BP 470/40 : DC 495 : EM BP 535/50
TRITC: EX BP 515/25 : DC 565 : EM BP 605/55
Texas Red: EX BP 560/20 : DC 595 : 630/60
Cy5: EX BP 620/60 : DC 660 : 700/75
Transmitted light modes: Bright-field and phase contrast
Objectives:
1x/0.04 NA (wd-3.2mm)
4x/0.2 NA (wd-20mm)
10x/0.45 NA (wd-4mm)
20x/0.75 NA (wd-1mm)
20x/0.45 NA ELWD Ph (wd- 6.9-8.2mm)
40x/0.95 NA (wd-0.25-0.17mm)
Software: Keyence View and Keyence Analyzer with dynamic cell count and measurement modules
Ownership: common
Location: Biomed 365B
Zeiss Live-Cell Imaging Widefield Microscope
Best suited for thin (less than 10 microns) samples mounted on a standard microscope slide with a coverslip, in a culture dish, or on a multi-well plate. Non-labeled and fluorescently-labeled samples may be used. Time course experiments requiring a temperature- or CO2-controlled environment or both.
Stand: inverted, Axiovert 200M
Stage: motorized stage for 3D stack and multi-point image acquisition; sample holder must be swapped to accept slides, petri-dishes, or multi-well plates.
Fluorescent filters available:
DAPI: EX 365 : DC 395 : EM BP 445/50
GFP: EX BP 470/40 : DC 495 : EM BP 525/50
TRITC: EX BP 540/25 : DC 565 : EM BP 605/55
Texas Red: EX BP 557.5/55 : DC 600 : EM LP 615
Objectives:
2.5x/0.075 NA (wd -8.7mm)
10x/0.25 NA (PH1) (wd-4.5mm)
20x/0.3 NA (PH1) (wd-4.3mm)
40x/0.6 NA (PH2) (DIC) (wd-0.75mm)
Software: Zeiss ZEN, Pecon Incubation Remote
Camera: Zeiss AxioCam MRm (monochrome)
Ownership: lab-owned
Location: 461 Biomedical Sciences
Please note that you must have documented BSLII clearance before you can use this scope.
Zeiss AxioImager Z2 Widefield Microscope
Best suited for samples up to ~40um (with Apotome) mounted on a standard microscope slide with a coverslip. Excels when extra-contrast is needed. Non-labeled, fluorescently-labeled and colorimetric-dye-labeled samples may be used.
Stand: upright, AxioImager Z2
Stage: motorized 8-slide stage for 3D stack and multi-point image acquisition
Fluorescent filters available:
DAPI: Fset49, EX 365 : DC 396 : EM BP 445/50
CFP: Fset47, EX BP 436/25 : DC 455 : EM BP 480/40
GFP: Fset38, EX BP 470/40 : DC 495 : EM BP 525/50
YFP: Fset46, EX BP 500/25 : DC 515 : EM BP 535/30
Cy3: Fset43, EX BP 550/25 : DC 570 : EM BP 605/70
RFP: Fset63, EX BP 572/25: DC 590 : EM BP 629/62
Cy5: Fset50, EX BP 640/30 : DC 660 : EM BP 690/50
Transmitted light modes: bright-field
Objectives:
2.5x/0.12 NA (wd-8.7mm)
10x/0.45 NA (wd-2mm)
20x/0.8 NA (wd-0.55mm)
40x/0.95 NA (wd-0.25mm)
40x/1.4 NA oil (wd-0.13mm)
63x/1.4 NA oil (wd-0.19mm)
100x/1.4 NA oil (wd-0.17mm)
Special Feature: Zeiss-Apotome 2
Software: Zeiss ZEN
Camera: Zeiss AxioCam 506 (monochrome) Zeiss AxioCam 506 (color)
Ownership: common
Location: 264 Biomedical Sciences
Please note that you must have documented BSLII clearance before you can use this scope.
M9 Multi-Focus Microscope (MFM)
This scope was built and designed by the Sara Abrahamsson Lab at UCSC. It captures 9 focal planes in the same instant through the use of diffractive optics. It is ideally suited for particle tracking in thin specimens with high temporal resolution. e.g. transcription loci.
Images are acquired in 9 focal planes within a 2.34 micron total volume. Each plane is spaced at 0.26 microns. This can be combined with a piezo drive based z-stack.
Laser excitation is at 488nm and/or 561nm. The dichroic is at 560nm. Green emission around 525 can be collected separately or simultaneously with a Red channel from 570-585nm.
Objective:
Olympus 60x/1.3 PLAN Apo Silicon Oil Immersion
Software: Micromanager for acquisition, MATLAB, FIJI and Visual Studio for processing and registration.
Camera: Hamamatsu Orca Fusion (Red channel), Andor iXon Ultra (Green channel)
Construction of this microscope was made possible through NSF Award ID 1828636.
Anyone seeking use of this instrument should contact Ben Abrams
Solamere Spinning Disk Confocal Microscope
Best suited for imaging of thick (10-150 microns) live samples mounted on a standard microscope slide with a coverslip, in a small dish, or in an incubation chamber. Non-labeled and fluorescently-labeled samples may be used.
Stand: inverted, Nikon TE2000
Stage: motorized stage for 3D stack and multipoint image acquisition; sample holder available to accept slides, petri-dishes, and multi-well plates
Fluorescent filters available:
For direct observation through eyepieces:
DAPI: EX BP 360/40 : DC 400 : EM BP 460/50
GFP: EX BP 470/40 : DC 495 : EM BP 525/50
TRITC: EX BP 540.5/25 : DC 565 : EM BP 620/60
For confocal operation:
DAPI: EM 460/50
GFP+RFP: EM BP 528/80
GFP: EM BP 525/50
RFP: EM BP 593/40
Cy5: EM BP 700/75
Transmitted light modes: bright-field and DIC
Laser lines: solid state 405, 488, 561, 640
Objectives:
10x/0.3 NA (wd-16mm)
20x/0.50 NA (DIC) (wd-not available)
40x/0.75 NA (wd-0.66mm)
40x/1.3 NA oil (wd-0.2mm)
60x/1.4 NA oil (DIC) (wd-0.13mm)
100x/1.4 NA oil (DIC) (wd-not available)
Software: Micro-Manager
Scanner: Yokogawa CSU-X1 spinning disk
Camera: Hamamatsu ImagEM X2
Ownership: common
Location: 111 Sinsheimer Labs
Leica SP5 Confocal Microscope
Best suited for thick (10-150 microns) samples mounted on a standard microscope slide with a coverslip or in a small petri dish. Used for FRAP, photo-activation, and scanning irregular regions of interest. Non-labeled and fluorescently-labeled samples may be used. Can capture highly dynamic processes if operated in resonance scanning mode.
Stand: inverted, Leica DMI6000
Stage: motorized stage for 3D stack, tiling, and multipoint image acquisition; sample holder will accommodate slides and small (60 mm) petri dishes
Fluorescent filters available:
For direct observation through eyepieces:
GFP
Texas Red
Simultaneous GFP + Texas Red
For confocal operation:
Spectral (AOTF)
Transmitted light modes: bright-field, DIC, and polarized light.
Laser lines: 405, Argon (458, 476, 488, 496, and 514) and HeNe (543, 594, and 633)
Objectives:
10x/0.3 NA (wd-11mm)
20x/0.75 NA (wd-0.59mm)
40x/1.25-0.75 NA oil DIC (wd-0.1mm)
63x/1.4-0.6 NA oil DIC (wd-0.1mm)
63x/1.2 NA water (wd-0.14mm)
Software: Leica Application Suite Advanced Fluorescence
Detectors: 3 channels available for simultaneous fluorescent image acquisition, 1 available for transmitted light image acquisition; standard scanning and fast resonance scanning (8000 Hz) modes available.
Ownership: common
Location: 111 Sinsheimer Labs
Zeiss 880 Confocal Microscope with Airyscan Fast
IMPORTANT:
If you publish data generated on this scope you must cite NIH S10 Grant 1S10OD23528-01.
Best suited for thick (10-150 microns) samples mounted on a standard microscope slide with a coverslip or in a petri-dish or multi-well plate. Non-labeled and fluorescently-labeled samples may be used. Time-course experiments on thick specimens requiring a temperature or CO2-controlled environment or both.
Stand: inverted, AxioObserver.Z1
Stage: motorized stage for 3D stack and multi-point image acquisition; sample holder available to accept slides, petri-dishes, and multi-well plates
Fluorescent filters available:
For direct observation through eyepieces:
DAPI
GFP
Rhodamine
mCherry
For confocal operation:
We have the “2+1” configuration which has 1 super-cooled multi-alkali, 1 standard multi-alkali and 1 GaAsP detector. These detectors are all spectral. The Airy detector can also be used in combination with these detectors to add a 4th channel that is filter based.
For Airyscan operation:
BP 420-480 + BP 495-550
BP 495-550 + LP570
BP 570-620 + LP645
BP 465-505 + LP 525
BP 420-480 + LP 605
Transmitted light modes: bright-field
Laser lines: 405, Argon (458, 488, and 514) and HeNe (561, 594 and 633)
Objectives:
10x/0.45 NA (wd-2.0mm)
20x/0.4 NA, corr LD (wd-8.4mm)
20x/0.8 NA (wd-0.55mm)
40x/0.95 NA corr (wd-0.25mm)
40x/1.2 water objective (wd-0.28mm)
63x/1.4 NA oil (wd-0.19mm)
Software: Zeiss ZEN Black for microscope control, ZEN Blue for additional processing
Detectors: 4 channels available for simultaneous fluorescent image acquisition, 1 channel available for transmitted light image acquisition
Ownership: common, but access must be approved by 880 confocal oversight committee until further notice. Please refer to the Zeiss 880 Confocal External User Policy about our access policy for this scope.
Location: 264 Biomedical Sciences
Please note that you must have documented BSLII clearance before you can use this scope.
Image Analysis Workstation
Best suited for post-acquisition processing and analysis of images where image processing software is operating-system-specific and where operations are RAM-intensive. Helps keep microscopes and image acquisition computers available for data acquisition.
Software:
- FIJI – Image J
- Zeiss LSM Browser
- Leica AF Lite Viewer
- Microsoft Excel
- Keyence Analyzer with Dynamic Cell Count and Measurement modules
- Keyence resizer
Ownership: common
Location: 111 Sinsheimer Labs
BioMed Image Analysis Workstation
Best suited for post-acquisition processing and analysis of images where image processing software is operating-system-specific and where operations are RAM-intensive. Helps keep microscopes and image acquisition computers available for data acquisition.
Software:
- Imaris
- AutoQuantX3
- Neurolucida
- Zeiss ZEN
- FIJI – Image J
- Zeiss LSM Browser
- Zeiss ZEN Lite
- Leica AF Lite Viewer
- Microsoft Office
- Keyence Analyzer with Dynamic Cell Count and Measurement modules
- Keyence resizer
Ownership: common
Location: 264 BioMed
Policies
Access
The facility is open to researchers inside and outside the life sciences community; however, investigators conducting life sciences research are given priority when reserving microscope time.
Permission and training from the microscopy center director is sufficient to use scopes owned in common. For lab-owned scopes, users must receive specific use permission.
Access to any equipment purchased on NIH instrumentation grants is subject to the terms of those grants and may require approval by the instrument advisory committee. Contact the center director if you wish to use a shared instrument in the facility or request services.
Training
Training and account set up is MANDATORY for all systems before they can be used independently. Please request training using the link above if you have not yet completed it. If it has been more than 6 months since you have used a particular microscope, before resuming use please contact the facility director for refresher training and updates. You are encouraged to reach out with questions at any time, regardless of how recently you trained.
Please see the Fee Schedule for questions regarding training fees.
General Use
- Be sure to read, understand an adhere to any policies posted by this facility.
- Be careful not to slop immersion oil around; it will create a sticky, dust-collecting mess. Clean the microscope stages and working area at the end of each section. Be sure to dispose of your used slides in the provided glass waste container. Spills or broken slides must be reported to the center manager immediately.
- Treat objectives with respect! Clean your slide before loading it on the stage; mixing different types of immersion oil may result in precipitation that is very difficult to clean.
- If you turn on the mercury lamp (for epi-fluorescence), it should stay on for at least 30 minutes. If you turn it off, it must stay off for 45 minutes before it can be turned on again.
- In the unlikely event of a mercury bulb blowing (with a loud pop sound), shut off the bulb power supply and evacuate the room of all users immediately. Put a “Stay Out” sign on the door and contact EH&S (831-459-2553) and the microscopy center manager (831-459-3999) immediately. The room must air out for at least 30 minutes.
- When you are done with your session, be sure to fill out the paper log next to the scope you are using.
- At the end of the session, place the objective turret in an empty or low mag position and place the stage in the appropriate position per scope- specific policy.
- Remember to place the dust cover back on the scope if you turn the scope off. Never place dust covers on the floor.
- If you experience any problems with a scope it is your responsibility to email the facility manager to report the problem as soon as possible (see also Communications Section).
Communication
Once you have been trained and have an active account you will be added to a scope-specific Google group to communicate issues specific to that scope with all other users of the instrument. This may include late changes in reservation status or issues with the hardware or software etc.
It is your responsibility to communicate problems with any scope or with the facility (e.g. missing supplies) with the facility director. You can do so by emailing the director directly or by emailing the scope-specific Google group.
If you need to contact another user directly, you can find out their email by clicking on their reservation in the reservation calendar and looking at the “Created by:” field. This will display their CruzID and you can add @ucsc.edu to the CruzID to get their email.
Facility Biohazard Procedures (BSL2)
All users must have an approved BUA on file and must fill out the safety questionnaire (see top of page) before doing any project in the facility.
Glove Policy: Gloves should only need to be worn while loading a hazardous sample onto the scope. Do not use soiled gloves on the microscope or computer controls.
People working in our Risk Group 2/BSL2 designated areas are also encouraged to take the “Biological Agents in Shared Facilities Awareness Training”, which is available through our campus Learning Center (login and “Find a Course” to search).
Be sure to disinfect any areas that might have become contaminated with a virulent agent during your session.
Scheduling
- All microscope sessions must be scheduled through the PBSci Equipment Reservation webpage. Log into the PBSci reservation webpage with your CruzID and password. If you are having trouble logging in to the PBSci site, please email help@ucsc.edu or call 831-459-4357, option 1.
- When making a reservation please enter your assigned microscopy user ID into the “Client’s name or Status:” field.
- Be realistic with your scheduling. The time that you schedule is time that someone else cannot use the equipment.
- It is your responsibility to know if someone has booked the scope after you. This policy may differ depending on which scope you are using, so be sure to obey the scope-specific guidelines provided during training. You should check the reservation web page no more than 30 minutes before the start of your session—please be sure that the web page has refreshed. Generally, if you are using a widefield microscope and the next user is coming 30 min. or less after your session ends, leave everything on (see additional note about LEDs below). If it will be more than 30 min before the next user, turn everything off. If you are using a confocal microscope and the next user is coming less than 2 hours after your session ends, leave everything on (see additional note about LEDs below), otherwise turn everything off—this will help maintain the health of lasers and other light sources.
- If you add a new reservation for a scope while another user is on that scope, or less than 30 minutes before the start of that user’s session, and your new reservation affects what they will do with the scope when they are done, it is your responsibility to tell that user and to ask them to leave the scope on for you.
- It is always OK to turn LED light sources off at the end of your session since they don’t require warm-up or cool down time.
- If you receive instrument-specific instructions about this during training, those instructions supersede the information in this policies section.
Cancellation
- If you must cancel a reservation please do so at least 24 hours before the scheduled session. If you finish early, please notify the next user.
- If you must cancel a reservation less than 24 hours before it is scheduled, please be considerate of other users and let them know by sending an email to the scope-specific Google group (mentioned above). Please adhere to the last-minute cancellation guidelines below.
- If you cancel your reservation less than 30 minutes before another person’s scheduled start time, it is your responsibility to make sure that the scope gets turned off after they are finished (they are expecting to leave it on)—either by turning the scope off yourself or by asking the previous user if he or she can do it for you.
- If you cancel your reservation less than 30 minutes before another user’s session is scheduled to end, it is your responsibility to turn the scope off.
Data Transfer and Disk Space
- The microscopy facility assumes NO responsibility for data storage. All hard drives will eventually fail. It is impossible to protect data from viruses with 100% confidence. It is the user’s responsibility to backup data.
- If you need to use the scope for data transfer to a portable hard drive, please reserve it as you would normally. This is the only way to ensure its availability. You should consider doing the data transfer while you are imaging to maximize scope availability and minimize your cost.
- It is recommended that you get a USB 3.0 drive (transfers ~10x faster than 2.0) or, even better, a USB-C drive, but this will require an adapter on most of our computers. It is recommended that you format the drive with ExFAT, this format is readable and writable by Windows and MacOS and has a larger than 4GB single file size limit.
- Users are encouraged to back up files frequently to reduce data transfer time.
- When saving data be sure to save it to the specified location on the specific computer that you are using – generally [Drive location] -> Image Data Folder -> Lab specific folder -> Your user folder.
- Effective December 1, 2021: User data must be removed from the acquisition computer within 90 days of collection. Data may need to be removed sooner if the drive gets full. Data that is older than 90 days may be deleted without further notice.
Citing the Facility and Instrumentation Grants
All publications containing research performed at the center must acknowledge the facility as: “Technical support from Benjamin Abrams, UCSC Life Sciences Microscopy Center, RRID: SCR_021135.”
Please alert the facility director when citing the facility so the publication can be recorded.
If you are using an instrument purchased with an instrumentation grant you must cite that grant number. See the instrument descriptions for specific information. This currently applies to users of the Zeiss 880 Confocal.
If you are publishing data generated on the 880 confocal please include the statement in your acknowledgements or methods section: “Purchase of the Zeiss 880 confocal microscope used in this research was made possible through the National Institutes of Health s10 Grant 1S10OD23528-01.”
Please refer to these additional guidelines about when acknowledgement vs. co-authorship is warranted. Please contact the IBSC if the Google drive link does not work.
Keys
Please use the links at the top of this page to request brass key access to Sinsheimer rooms or key card access to BioMed. Please be sure to list your P.I. as your supervisor on your key requests, not the Microscopy Center Director.
Food and Drink
No eating or drinking is permitted in the microscope rooms.
Publications
If you have a recent publication that acknowledges the UCSC Life Sciences Microscopy Center that is not listed below, please contact us and provide the PMID for your publication.
- Warecki B, Bast I, Tajima M, Sullivan W. Connections between sister and non-sister telomeres of segregating chromatids maintain euploidy. Current Biology. 2023;33(1):58-74. e5.
- Monem PC, Vidyasagar N, Piatt AL, Sehgal E, Arribere JA. Ubiquitination of stalled ribosomes enables mRNA decay via HBS-1 and NONU-1 in vivo. PLoS Genet. 2023;19(1):e1010577. Epub 20230110. doi: 10.1371/journal.pgen.1010577. PubMed PMID: 36626369; PMCID: PMC9870110.
- Abrams B, Pengo T, Wee T-L, Deagle RC, Vuillemin N, Callahan LM, Smith MA, Kubow KE, Girard A-M, Rappoport JZ, Bayles CJ, Cameron LA, Cole R, Brown CM. Tissue-Like 3D Standard and Protocols for Microscope Quality Management. Microscopy and Microanalysis. 2023. doi: 10.1093/micmic/ozad014.
- Warecki B, Titen SWA, Alam MS, Vega G, Lemseffer N, Hug K, Minden JS, Sullivan W. Wolbachia action in the sperm produces developmentally deferred chromosome segregation defects during the Drosophila mid-blastula transition. Elife. 2022;11. Epub 20220923. doi: 10.7554/eLife.81292. PubMed PMID: 36149408; PMCID: PMC9507124.
- Tsyporin J, Tastad D, Ma X, Nehme A, Finn T, Huebner L, Liu G, Gallardo D, Makhamreh A, Roberts JM. Transcriptional repression by FEZF2 restricts alternative identities of cortical projection neurons. Cell reports. 2022;35(12):109269.
- Sikandar SS, Gulati GS, Antony J, Fetter I, Kuo AH, Ho WHD, Haro-Acosta V, Das S, Steen CB, Pereira TA. Identification of a minority population of LMO2+ breast cancer cells that integrate into the vasculature and initiate metastasis. Science Advances. 2022;8(45):eabm3548.
- Seiler ST, Mantalas GL, Selberg J, Cordero S, Torres-Montoya S, Baudin PV, Ly VT, Amend F, Tran L, Hoffman RN, Rolandi M, Green RE, Haussler D, Salama SR, Teodorescu M. Modular automated microfluidic cell culture platform reduces glycolytic stress in cerebral cortex organoids. Sci Rep. 2022;12(1):20173. Epub 20221123. doi: 10.1038/s41598-022-20096-9. PubMed PMID: 36418910; PMCID: PMC9684529.
- Ricemeyer L, Aguilar-Hernández N, López T, Espinosa R, Lanning S, Mukherjee S, Cuellar C, López S, Arias CF, DuBois RM. Structures of two human astrovirus capsid/neutralizing antibody complexes reveal distinct epitopes and inhibition of virus attachment to cells. Journal of virology. 2022;96(1):e01415-21.
- Palmer J, Segura CJ, Matsushima L, Abrams B, Lee HW, Ayzner AL. Conjugated polyelectrolyte-based ternary exciton funnels via liposome scaffolds. Mol Syst Des Eng. 2022;7(4):392-402. doi: 10.1039/d1me00139f. PubMed PMID: WOS:000749246400001.
- Johnston AR, Pitch GM, Minckler ED, Mora IG, Balasco Serrao VH, Dailing EA, Ayzner AL. Excitonically Coupled Simple Coacervates via Liquid/Liquid Phase Separation. J Phys Chem Lett. 2022;13(44):10275-81. Epub 20221028. doi: 10.1021/acs.jpclett.2c02466. PubMed PMID: 36305559; PMCID: PMC9661528.
- Johnston AR, Minckler ED, Shockley MC, Matsushima LN, Perry SL, Ayzner AL. Conjugated Polyelectrolyte‐Based Complex Fluids as Aqueous Exciton Transport Networks. Angewandte Chemie. 2022;134(20):e202117759.
- Johnston AR. Investigation into the Photophysics and Phase Behavior of Conjugated Polyelectrolyte Complexes: UC Santa Cruz; 2022.
- Bowman B. The Structure of Prevacuolar Compartments in Neurospora Crassa as Observed with Super-Resolution Microscopy. Available at SSRN 4221732. 2022.
- Abed S, Reilly A, Arnold SJ, Feldheim DA. Adult expression of Tbr2 is required for the maintenance but not survival of intrinsically photosensitive retinal ganglion cells. Frontiers in Cellular Neuroscience. 2022;16.
- Smith-Berdan S, Bercasio A, Kramer L, Petkus B, Hinck L, Forsberg EC. Acute and endothelial-specific Robo4 deletion affect hematopoietic stem cell trafficking independent of VCAM1. PLoS One. 2021;16(8):e0255606. Epub 20210813. doi: 10.1371/journal.pone.0255606. PubMed PMID: 34388149; PMCID: PMC8362960.
- Perna J, Lu J, Mullen B, Liu T, Tjia M, Weiser S, Ackman J, Zuo Y. Perinatal penicillin exposure affects cortical development and sensory processing. Frontiers in Molecular Neuroscience. 2021:323.
- Cazares O, Chatterjee S, Lee P, Strietzel C, Bubolz JW, Harburg G, Howard J, Katzman S, Sanford J, Hinck L. Alveolar progenitor differentiation and lactation depends on paracrine inhibition of notch via ROBO1/CTNNB1/JAG1. Development. 2021;148(21). Epub 20211110. doi: 10.1242/dev.199940. PubMed PMID: 34758082; PMCID: PMC8627605.
- Wu DC, Zamorano-Sanchez D, Pagliai FA, Park JH, Floyd KA, Lee CK, Kitts G, Rose CB, Bilotta EM, Wong GCL, Yildiz FH. Reciprocal c-di-GMP signaling: Incomplete flagellum biogenesis triggers c-di-GMP signaling pathways that promote biofilm formation. PLoS Genet. 2020;16(3):e1008703. Epub 20200316. doi: 10.1371/journal.pgen.1008703. PubMed PMID: 32176702; PMCID: PMC7098655.
- Warecki B, Ling X, Bast I, Sullivan W. ESCRT-III–mediated membrane fusion drives chromosome fragments through nuclear envelope channels. Journal of Cell Biology. 2020;219(3).
- Schwechheimer C, Hebert K, Tripathi S, Singh PK, Floyd KA, Brown ER, Porcella ME, Osorio J, Kiblen JT, Pagliai FA. A tyrosine phosphoregulatory system controls exopolysaccharide biosynthesis and biofilm formation in Vibrio cholerae. PLoS Pathogens. 2020;16(8):e1008745.
- Joshi A, Mahmoud SA, Kim SK, Ogdahl JL, Lee VT, Chien P, Yildiz FH. c-di-GMP inhibits LonA-dependent proteolysis of TfoY in Vibrio cholerae. PLoS Genet. 2020;16(6):e1008897. Epub 20200626. doi: 10.1371/journal.pgen.1008897. PubMed PMID: 32589664; PMCID: PMC7371385.
- Jasani A, Huynh T, Kellogg DR. Growth-dependent activation of protein kinases suggests a mechanism for measuring cell growth. Genetics. 2020;215(3):729-46.
- Hathroubi S, Zerebinski J, Clarke A, Ottemann KM. Helicobacter pylori biofilm confers antibiotic tolerance in part via a protein-dependent mechanism. Antibiotics. 2020;9(6):355.
- Hathroubi S, Hu S, Ottemann KM. Genetic requirements and transcriptomics of Helicobacter pylori biofilm formation on abiotic and biotic surfaces. npj Biofilms and Microbiomes. 2020;6(1):56.
- Glover ML, Burroughs AM, Monem PC, Egelhofer TA, Pule MN, Aravind L, Arribere JA. NONU-1 Encodes a Conserved Endonuclease Required for mRNA Translation Surveillance. Cell Rep. 2020;30(13):4321-31 e4. doi: 10.1016/j.celrep.2020.03.023. PubMed PMID: 32234470; PMCID: PMC7184879.
- Gallego-Hernandez A, DePas W, Park J, Teschler J, Hartmann R, Jeckel H, Drescher K, Beyhan S, Newman D, Yildiz F. Upregulation of virulence genes promotes Vibrio cholerae biofilm hyperinfectivity. Proceedings of the National Academy of Sciences. 2020;117(20):11010-7.
- Foley AR, Lee HW, Raskatov JA. A Focused Chiral Mutant Library of the Amyloid beta 42 Central Electrostatic Cluster as a Tool To Stabilize Aggregation Intermediates. J Org Chem. 2020;85(3):1385-91. Epub 20200103. doi: 10.1021/acs.joc.9b02312. PubMed PMID: 31875394.
- Floyd KA, Lee CK, Xian W, Nametalla M, Valentine A, Crair B, Zhu S, Hughes HQ, Chlebek JL, Wu DC, Hwan Park J, Farhat AM, Lomba CJ, Ellison CK, Brun YV, Campos-Gomez J, Dalia AB, Liu J, Biais N, Wong GCL, Yildiz FH. c-di-GMP modulates type IV MSHA pilus retraction and surface attachment in Vibrio cholerae. Nat Commun. 2020;11(1):1549. Epub 20200325. doi: 10.1038/s41467-020-15331-8. PubMed PMID: 32214098; PMCID: PMC7096442.
- Leitao RM, Jasani A, Talavara RA, Pham A, Okobi QJ, Kellogg DR. A Conserved PP2A Regulatory Subunit Enforces Proportional Relationships Between Cell Size and Growth Rate. Genetics. 2019;213(2):517-28. Epub 20190905. doi: 10.1534/genetics.119.301012. PubMed PMID: 31488515; PMCID: PMC6781898.
- Kitts G, Giglio KM, Zamorano-Sánchez D, Park JH, Townsley L, Cooley RB, Wucher BR, Klose KE, Nadell CD, Yildiz FH. A conserved regulatory circuit controls large adhesins in Vibrio cholerae. MBio. 2019;10(6):e02822-19.
- Foley AR, Finn TS, Kung T, Hatami A, Lee H-W, Jia M, Rolandi M, Raskatov JA. Trapping and characterization of nontoxic Aβ42 aggregation intermediates. ACS Chemical Neuroscience. 2019;10(8):3880-7.
- Field AR, Jacobs FM, Fiddes IT, Phillips AP, Reyes-Ortiz AM, LaMontagne E, Whitehead L, Meng V, Rosenkrantz JL, Olsen M. Structurally conserved primate LncRNAs are transiently expressed during human cortical differentiation and influence cell-type-specific genes. Stem Cell Reports. 2019;12(2):245-57.
- Cornejo‐Castillo FM, Muñoz‐Marín MdC, Turk‐Kubo KA, Royo‐Llonch M, Farnelid H, Acinas SG, Zehr JP. UCYN‐A3, a newly characterized open ocean sublineage of the symbiotic N2‐fixing cyanobacterium Candidatus Atelocyanobacterium thalassa. Environmental Microbiology. 2019;21(1):111-24.
- Moyer CE, Hiolski EM, Marcinek DJ, Lefebvre KA, Smith DR, Zuo Y. Repeated low level domoic acid exposure increases CA1 VGluT1 levels, but not bouton density, VGluT2 or VGAT levels in the hippocampus of adult mice. Harmful algae. 2018;79:74-86.
- Morgan JM, Lam HN, Delgado J, Luu J, Mohammadi S, Isberg RR, Wang H, Auerbuch V. An Experimental Pipeline for Initial Characterization of Bacterial Type III Secretion System Inhibitor Mode of Action Using Enteropathogenic Yersinia. Front Cell Infect Microbiol. 2018;8:404. Epub 20181122. doi: 10.3389/fcimb.2018.00404. PubMed PMID: 30524970; PMCID: PMC6262202.
- Fiddes IT, Lodewijk GA, Mooring M, Bosworth CM, Ewing AD, Mantalas GL, Novak AM, van den Bout A, Bishara A, Rosenkrantz JL. Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell. 2018;173(6):1356-69. e22.
- Byrne G, O’Rourke SM, Alexander DL, Yu B, Doran RC, Wright M, Chen Q, Azadi P, Berman PW. CRISPR/Cas9 gene editing for the creation of an MGAT1-deficient CHO cell line to control HIV-1 vaccine glycosylation. PLoS Biol. 2018;16(8):e2005817. Epub 20180829. doi: 10.1371/journal.pbio.2005817. PubMed PMID: 30157178; PMCID: PMC6133382.
- Vickers ET, Garai M, Bonabi Naghadeh S, Lindley S, Hibbs J, Xu Q-H, Zhang JZ. Two-photon photoluminescence and photothermal properties of hollow gold nanospheres for efficient theranostic applications. The Journal of Physical Chemistry C. 2017;122(25):13304-13.
- Tjia M, Yu X, Jammu LS, Lu J, Zuo Y. Pyramidal neurons in different cortical layers exhibit distinct dynamics and plasticity of apical dendritic spines. Frontiers in neural circuits. 2017;11:43.
- Teschler JK, Cheng AT, Yildiz FH. The two-component signal transduction system VxrAB positively regulates Vibrio cholerae biofilm formation. Journal of bacteriology. 2017;199(18):e00139-17.
- Tao X, Lin H-H, Lam T, Rodriguez R, Wang JW, Kubby J. Transcutical imaging with cellular and subcellular resolution. Biomedical optics express. 2017;8(3):1277-89.
- Pisano F, Zampaglione E, McAlinden N, Roebber J, Dawson MD, Mathieson K, Sher A. Large scale matching of function to the genetic identity of retinal ganglion cells. Sci Rep. 2017;7(1):15395. Epub 20171113. doi: 10.1038/s41598-017-15741-7. PubMed PMID: 29133846; PMCID: PMC5684394.
- Ozcelik D, Jain A, Stambaugh A, Stott MA, Parks JW, Hawkins A, Schmidt H. Scalable Spatial-Spectral Multiplexing of Single-Virus Detection Using Multimode Interference Waveguides. Sci Rep. 2017;7(1):12199. Epub 20170922. doi: 10.1038/s41598-017-12487-0. PubMed PMID: 28939852; PMCID: PMC5610187.
- Morgan JM, Duncan MC, Johnson KS, Diepold A, Lam H, Dupzyk AJ, Martin LR, Wong WR, Armitage JP, Linington RG, Auerbuch V. Piericidin A1 Blocks Yersinia Ysc Type III Secretion System Needle Assembly. mSphere. 2017;2(1). Epub 20170215. doi: 10.1128/mSphere.00030-17. PubMed PMID: 28217742; PMCID: PMC5311113.
- Luo Q, Ding J, Tao X, Lu J, Lam T, Rodriguez R, Zuo Y, Kubby J. A three-photon microscope with adaptive optics for deep-tissue in vivo structural and functional brain imaging. Neural Imaging and Sensing2017.
- Leitao RM, Kellogg DR. The duration of mitosis and daughter cell size are modulated by nutrients in budding yeast. Journal of Cell Biology. 2017;216(11):3463-70.
- Lam H, Schwochert J, Lao Y, Lau T, Lloyd C, Luu J, Kooner O, Morgan J, Lokey S, Auerbuch V. Synthetic Cyclic Peptomers as Type III Secretion System Inhibitors. Antimicrob Agents Chemother. 2017;61(9). Epub 20170824. doi: 10.1128/AAC.00060-17. PubMed PMID: 28652236; PMCID: PMC5571333.
- Knutson AKa, Egelhofer T, Rechtsteiner A, Strome S. Germ granules prevent accumulation of somatic transcripts in the adult Caenorhabditis elegans germline. Genetics. 2017;206(1):163-78.
- Hodges JL, Yu X, Gilmore A, Bennett H, Tjia M, Perna JF, Chen CC, Li X, Lu J, Zuo Y. Astrocytic Contributions to Synaptic and Learning Abnormalities in a Mouse Model of Fragile X Syndrome. Biol Psychiatry. 2017;82(2):139-49. Epub 20160913. doi: 10.1016/j.biopsych.2016.08.036. PubMed PMID: 27865451; PMCID: PMC5348290.
- Fong JC, Rogers A, Michael AK, Parsley NC, Cornell WC, Lin YC, Singh PK, Hartmann R, Drescher K, Vinogradov E, Dietrich LE, Partch CL, Yildiz FH. Structural dynamics of RbmA governs plasticity of Vibrio cholerae biofilms. Elife. 2017;6. Epub 20170801. doi: 10.7554/eLife.26163. PubMed PMID: 28762945; PMCID: PMC5605196.
- Duncan MC, Herrera NG, Johnson KS, Engel JN, Auerbuch V. Bacterial internalization is required to trigger NIK-dependent NF-kappaB activation in response to the bacterial type three secretion system. PLoS One. 2017;12(2):e0171406. Epub 20170206. doi: 10.1371/journal.pone.0171406. PubMed PMID: 28166267; PMCID: PMC5293232.
- Brose L, Crest J, Tao L, Sullivan W. Polo kinase mediates the phosphorylation and cellular localization of Nuf/FIP3, a Rab11 effector. Molecular biology of the cell. 2017;28(11):1435-43.
- Beier C, Hovhannisyan A, Weiser S, Kung J, Lee S, Lee DY, Huie P, Dalal R, Palanker D, Sher A. Deafferented adult rod bipolar cells create new synapses with photoreceptors to restore vision. Journal of Neuroscience. 2017;37(17):4635-44.
- Rogers A, Townsley L, Gallego-Hernandez AL, Beyhan S, Kwuan L, Yildiz FH. The LonA Protease Regulates Biofilm Formation, Motility, Virulence, and the Type VI Secretion System in Vibrio cholerae. J Bacteriol. 2016;198(6):973-85. Epub 20160111. doi: 10.1128/JB.00741-15. PubMed PMID: 26755629; PMCID: PMC4772603.
- Miller HK, Schwiesow L, Au-Yeung W, Auerbuch V. Hereditary hemochromatosis predisposes mice to Yersinia pseudotuberculosis infection even in the absence of the type III secretion system. Frontiers in cellular and infection microbiology. 2016;6:69.
- Le LT, Cazares O, Mouw JK, Chatterjee S, Macias H, Moran A, Ramos J, Keely PJ, Weaver VM, Hinck L. Loss of miR-203 regulates cell shape and matrix adhesion through ROBO1/Rac/FAK in response to stiffness. J Cell Biol. 2016;212(6):707-19. doi: 10.1083/jcb.201507054. PubMed PMID: 26975850; PMCID: PMC4792073.
- Farnelid H, Turk-Kubo K, Zehr J. Identification of associations between bacterioplankton and photosynthetic picoeukaryotes in coastal waters. Front Microbiol. 2016; 7: 1–16. 2016.
- Farnelid H, Turk-Kubo K, Muñoz-Marín MC, Zehr JP. New insights into the ecology of the globally significant uncultured nitrogen-fixing symbiont UCYN-A. Aquatic Microbial Ecology. 2016;77(3):125-38. doi: 10.3354/ame01794.
- Cheng TL, Mayberry H, McGuire LP, Hoyt JR, Langwig KE, Nguyen H, Parise KL, Foster JT, Willis CKR, Kilpatrick AM, Frick WF. Efficacy of a probiotic bacterium to treat bats affected by the disease white‐nose syndrome. Journal of Applied Ecology. 2016;54(3):701-8. doi: 10.1111/1365-2664.12757.
- Roelofs KG, Jones CJ, Helman SR, Shang X, Orr MW, Goodson JR, Galperin MY, Yildiz FH, Lee VT. Systematic Identification of Cyclic-di-GMP Binding Proteins in Vibrio cholerae Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems. PLoS Pathog. 2015;11(10):e1005232. Epub 20151027. doi: 10.1371/journal.ppat.1005232. PubMed PMID: 26506097; PMCID: PMC4624772.
- Ozel RE, Lohith A, Mak WH, Pourmand N. Single-cell intracellular nano-pH probes. RSC Adv. 2015;5(65):52436-43. Epub 20150609. doi: 10.1039/c5ra06721a. PubMed PMID: 27708772; PMCID: PMC5047438.
- Karg T, Warecki B, Sullivan W. Aurora B-mediated localized delays in nuclear envelope formation facilitate inclusion of late-segregating chromosome fragments. Mol Biol Cell. 2015;26(12):2227-41. Epub 20150415. doi: 10.1091/mbc.E15-01-0026. PubMed PMID: 25877868; PMCID: PMC4462941.
- Jones CJ, Utada A, Davis KR, Thongsomboon W, Zamorano Sanchez D, Banakar V, Cegelski L, Wong GC, Yildiz FH. C-di-GMP regulates motile to sessile transition by modulating MshA pili biogenesis and near-surface motility behavior in Vibrio cholerae. PLoS pathogens. 2015;11(10):e1005068.
- Eckler MJ, Nguyen TD, McKenna WL, Fastow BL, Guo C, Rubenstein JL, Chen B. Cux2-positive radial glial cells generate diverse subtypes of neocortical projection neurons and macroglia. Neuron. 2015;86(4):1100-8.
- Bowman BJ, Draskovic M, Schnittker RR, El-Mellouki T, Plamann MD, Sánchez-León E, Riquelme M, Bowman EJ. Characterization of a novel prevacuolar compartment in Neurospora crassa. Eukaryotic cell. 2015;14(12):1253-63.
- Bilecen K, Fong JC, Cheng A, Jones CJ, Zamorano-Sánchez D, Yildiz FH. Polymyxin B resistance and biofilm formation in Vibrio cholerae are controlled by the response regulator CarR. Infection and immunity. 2015;83(3):1199-209.
- Miller HK, Kwuan L, Schwiesow L, Bernick DL, Mettert E, Ramirez HA, Ragle JM, Chan PP, Kiley PJ, Lowe TM, Auerbuch V. IscR is essential for yersinia pseudotuberculosis type III secretion and virulence. PLoS Pathog. 2014;10(6):e1004194. Epub 20140612. doi: 10.1371/journal.ppat.1004194. PubMed PMID: 24945271; PMCID: PMC4055776.
- Harburg G, Compton J, Liu W, Iwai N, Zada S, Marlow R, Strickland P, Zeng YA, Hinck L. SLIT/ROBO2 signaling promotes mammary stem cell senescence by inhibiting Wnt signaling. Stem cell reports. 2014;3(3):385-93.
- Effenberger KA, Anderson DD, Bray WM, Prichard BE, Ma N, Adams MS, Ghosh AK, Jurica MS. Coherence between cellular responses and in vitro splicing inhibition for the anti-tumor drug pladienolide B and its analogs. Journal of Biological Chemistry. 2014;289(4):1938-47.
- Eckler MJ, Larkin KA, McKenna WL, Katzman S, Guo C, Roque R, Visel A, Rubenstein JL, Chen B. Multiple conserved regulatory domains promote Fezf2 expression in the developing cerebral cortex. Neural development. 2014;9:1-12.
- Yu X, Wang G, Gilmore A, Yee AX, Li X, Xu T, Smith SJ, Chen L, Zuo Y. Accelerated experience-dependent pruning of cortical synapses in ephrin-A2 knockout mice. Neuron. 2013;80(1):64-71. Epub 20131002. doi: 10.1016/j.neuron.2013.07.014. PubMed PMID: 24094103; PMCID: PMC3792401.
- Masuda M, Braun-Sommargren M, Crooks D, Smith DR. Golgi phosphoprotein 4 (GPP130) is a sensitive and selective cellular target of manganese exposure. Synapse. 2013;67(5):205-15. Epub 20130208. doi: 10.1002/syn.21632. PubMed PMID: 23280773; PMCID: PMC3987769.
- Guo C, Eckler MJ, McKenna WL, McKinsey GL, Rubenstein JL, Chen B. Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron. 2013;80(5):1167-74.
- Tao X, Crest J, Kotadia S, Azucena O, Chen DC, Sullivan W, Kubby J. Live imaging using adaptive optics with fluorescent protein guide-stars. Optics express. 2012;20(14):15969-82.
- Fasulo B, Deuring R, Murawska M, Gause M, Dorighi KM, Schaaf CA, Dorsett D, Brehm A, Tamkun JW. The Drosophila MI-2 chromatin-remodeling factor regulates higher-order chromatin structure and cohesin dynamics in vivo2012.
Funding/Grant Acknowledgement
The UCSC Life Sciences Microscopy Center is a core facility supported by the UCSC Division of Physical and Biological Sciences and the California Institute for Quantitative Biosciences (QB3). All publications, posters, presentations, etc. that utilize the UCSC Life Sciences Microscopy Center must acknowledge RRID SCR_021135.
Contact
Ben Abrams, Director
Email: babrams@ucsc.edu
Phone: (831) 459-3999
Office: 150 Sinsheimer Labs
